Deep topographic proteomics of a human brain tumour.
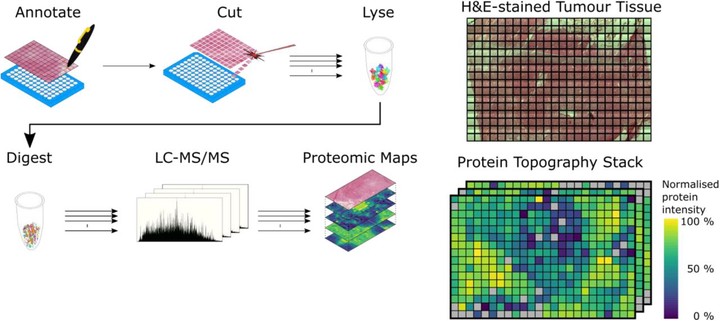 Image credit: Simon Davis
Image credit: Simon DavisAbstract:
Cellular protein expression profiles within tissues are key to understanding disease pathology, and their spatial organisation determines cellular function. To precisely define molecular phenotypes in the spatial context of tissue, there is a need for unbiased, quantitative technology capable of mapping the expression of many hundreds to thousands of proteins within tissue structures. Here, we present a workflow for spatially resolved, quantitative proteomics of tissue that generates maps of protein expression across a tissue slice derived from a human atypical teratoid-rhabdoid tumour (AT/RT). We employ spatially-aware statistical methods that do not require prior knowledge of tissue structure to highlight proteins and pathways with varying spatial abundance patterns. We identify novel aspects of AT/RT biology that map onto the brain-tumour interface. Overall, this work informs on methods for spatially resolved deep proteo-phenotyping of tissue heterogeneity. Advanced spatially resolved tissue proteomics will push the boundaries of understanding tissue biology and pathology at the molecular level.