Spatially resolved MALDI mass spectrometric imaging of human brain tissue for metabolites, lipids, and peptides - proof of principle
Introduction: Matrix-assisted laser desorption/ionization (MALDI) mass spectrometric imaging (MALDI-MSI) can be used to image the distribution of individual biomolecules within tissue sections. This can provide unique insights into the spatial relationship between peptides, lipids, and small molecules and potentially highlight underlying metabolic pathways. Until recently the limited resolution of MALDI MSI made it challenging to extrapolate biochemical changes to single-cell populations. In this proof of principle experiment we used highly sensitive, high-resolution MALDI MSI to quantitatively measure and compare the intensities of peptides, lipids and small molecules in the motor cortex and cerebellum from healthy people and those with amyotrophic lateral sclerosis (ALS).
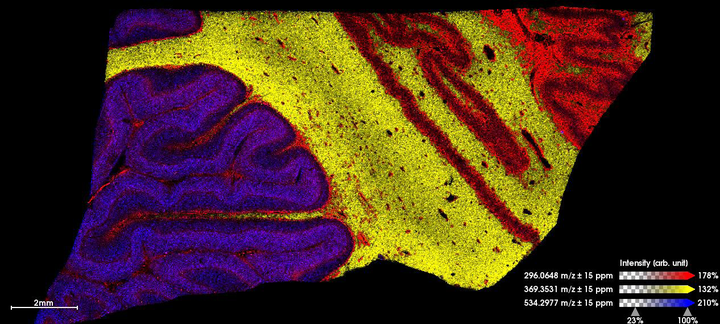
Methodology: The primary motor cortex and cerebellum from a healthy donor and a brain with genetic C9orf72-associated ALS was selected from the Oxford Brain Bank. A systematic sectioning protocol was used to align downstream proteomic, lipidomic, and metabolic MALDI MSI findings with standard neuropathology and laser microdissected (LCM) LC-MS/MS. Sections destined for MALDI imaging were dried in a vacuum and sent to Bruker Daltonik to be analysed on their tims-TOF flex mass spectrometer. Samples were analysed with and without the novel MALDI-2 attachment. Lipids and small molecules were imaged at a resolution of 20µm and peptides were imaged at 50µm.
Results: In this proof of principle experiment, we used receiver operating characteristic (ROC) analysis and observed striking global differences in the abundance of peptides, lipids, and small molecules in compartmentalised areas of the human brain. The granular layer of the cerebellum and the grey matter of the motor cortex are known to contain neurons that are selectively vulnerable to C9orf72-muation-driven ALS pathology. Impressively, the resolution capabilities of the tims-TOF flex (MALDI-2) mode made it possible to resolve histological features not visualised by traditional MALDI MSI e.g. individual neuronal layers, and the magnocellular neurons of the cerebellum and motor cortex, principally the Purkinje cell, Betz cell and the cells of the Dentate Nucleus. MALDI-informed LCM-directed mass spectrometry is currently underway to validate the identity of these differentially expressed biomolecules in individual cell populations.
Conclusion: In this proof of principle experiment, we demonstrate that MALDI-MSI combined with classical LCM LC-MS/MS has the potential to spatially resolve metabolic signatures in human brain at cellular resolution.