The microscopy connectome towards 3D PLI tractography in the BigMac dataset
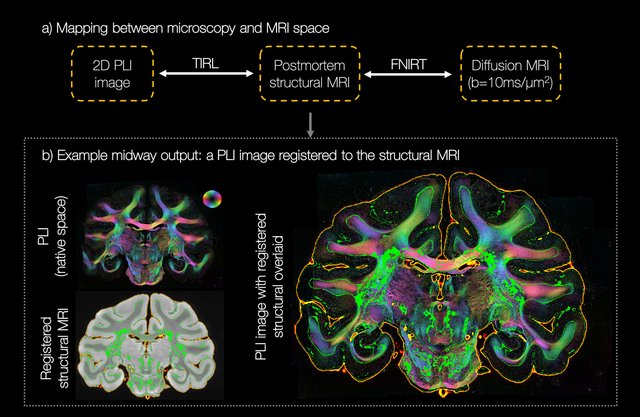 Image credit: Amy Howard
Image credit: Amy HowardIntroduction:
Multimodal datasets, which combine diffusion MRI (dMRI) and microscopy data that offers a ‘ground truth’ estimate of the connectome, provide a unique opportunity to both assess the performance of tractography and, crucially, drive algorithm design. To facilitate fair comparison, the multimodal data is ideally i) acquired in the same single brain with ii) whole brain coverage, iii) the microscopy and MRI data are co-registered together with high precision and iv) the microscopy is informative of the 3D connectome and not limited to 2D analysis (as is typical with many 2D microscopy techniques).
The BigMac dataset 1 is a novel resource that has the potential to fulfil these criteria. The BigMac dataset combines extensive MRI data (both in vivo and postmortem) with multi- contrast microscopy acquired throughout a single, macaque brain (Figure 1, criteria i&ii). In this work we demonstrate precise co-registration of the MRI and microscopy data (criterion iii), and reconstruct the 2D microscopy fibre orientations as full 3D vectors in MRI space (criterion iv). This will facilitate future whole brain reconstruction of the 3D microscopy connectome at ultra-high spatial resolution. We expect this microscopy connectome to both provide new anatomical insight, and be a valuable resource for the diffusion modelling community. Upon publication, the BigMac data and tools will be made openly available.
Methods:
The BigMac dataset includes polarised light imaging 2,3,4 data (PLI, Figure 1 bottom) that estimates the primary fibre orientation of myelinated fibres per microscopy pixel. In BigMac, Coronal PLI data were acquired every 350 μm along the rostro-caudal axis, with an in-plane resolution of 4 μm per pixel. Here we take the PLI estimate of the fibre orientations within the microscopy plane, and approximate the through-plane ‘inclination’ angle with that from dMRI (the ball and stick model 5,6 ), to reconstruct 3D ‘hybrid dMRI-microscopy fibre orientations’ at the resolution of the microscopy data.
Diffusion MRI data (b=10ms/μm 2 , 1000 gradient directions, 1mm isotropic) were analysed using the ball and stick 5,6 (BAS) model to estimate fibre populations per voxel, with 50 fibre orientation estimates or ‘samples’ per population. The PLI images were co-registered to the dMRI data using an optimised TIRL 7 protocol (Figure 2). The in-plane angle was warped into the diffusion space (accounting for any necessary rotations) and compared to the BAS samples within the corresponding voxel. To facilitate fair comparison, the BAS samples were projected onto the microscopy plane. Samples from BAS fibre populations with signal fractions <0.1 were excluded. Finally, the microscopy through-plane angle was approximated by that from the most similar BAS sample. This produced a hybrid dMRI-microscopy 3D fibre orientation per microscopy pixel. To have fibre orientations populate a substantial volume in dMRI space, the process was repeated for 30 consecutive PLI sections. The pixelwise fibre orientations were then combined into 3D fibre orientation distributions (FODs) for comparison with dMRI equivalents. Here, a set of voxels were defined in dMRI space at some specified resolution. In each voxel, the hybrid dMRI-microscopy fibre orientations populated a 3D ‘frequency histogram’ defined by 256 points evenly spaced across the sphere. Spherical harmonics of order 8 were then fitted to the normalised histogram. Because these FODs were output as spherical harmonic coefficients, the hybrid dMRI-microscopy FODs could then be visualised in standard MRI viewers 9,10 and input into existing tractography methods 10 .
For comparison, the dMRI data were also processed using constrained spherical deconvolution (CSD) 8.
Results:
Figure 2 shows excellent registration of the 2D PLI images into the 3D MRI volume, with close alignment of the tissue boundaries.
In our method, the through-plane microscopy orientation is approximated by that from the BAS model. This assumption is most valid when the PLI and BAS fibre orientations in the microscopy plane are highly similar, as demonstrated throughout the majority of the white matter (Figure 3 top).
Figure 3 bottom shows example hybrid dMRI-PLI 3D fibre orientations from a single microscopy section. Near the microscopy plane (LR-IS), we retain high resolution information from the microscopy data (4x4 μm). The through-plane information is of coarser resolution, comparative to the MRI data.
The hybrid dMRI-PLI fibre orientations were then reconstructed into FODs at varying spatial resolutions (Figures 4&5). Reassuringly, the hybrid dMRI-microscopy FODs show smoothly varying patterns in all three dimensions, but notably reconstruct fewer crossing fibre populations than that suggested by dMRI (Figure 4). Furthermore, we observe smoothly varying through-plane fibre orientations even when the hybrid FODs are reconstructed at a higher spatial resolution than the dMRI data (0.6 versus 1 mm, Figure 5). Finally, the hybrid FODs can be reconstructed at very high in-plane resolutions, up to the native microscopy resolution (4x4 μm in-plane, 350 μm though-plane).
References:
- Howard AFD, et al. The BigMac dataset: ultra-high angular resolution diffusion imaging and multi-contrast microscopy of a whole macaque brain, ISMRM 27th Annual Meeting, 2019.
- Axer H, et al. Quantitative estimation of 3-D fiber course in gross histological sections of the human brain using polarized light, Journal of Neuroscience Methods, 2001
- Larsen L, et al. Polarized light imaging of white matter architecture, Microscopy Research and Technique, 2007
- Axer M, et al. High-Resolution Fiber Tract Reconstruction in the Human Brain by Means of Three-Dimensional Polarized Light Imaging, Frontiers in Neuroinformatics, 2011
- Behrens TE, et al. Characterization and propagation of uncertainty in diffusion-weighted MR imaging. Magnetic Resonance in Medicine, 2003
- Behrens TE, et al. Probabilistic diffusion tractography with multiple fibre orientations. What can we gain? NeuroImage , 2007.
- Huszar IN, et al. Tensor Image Registration Library: Automated Non-Linear Registration of Sparsely Sampled Histological Specimens to Post-Mortem MRI of the Whole Human Brain, bioRxiv, 2019
- Tournier JD, et al. Robust determination of the fibre orientation distribution in diffusion MRI: non-negativity constrained super-resolved spherical deconvolution. Neuroimage, 2007
- McCarthy P, FSLeyes, Zenodo, 2020
- Tournier JD, et al. MRtrix3: A fast, flexible and open software framework for medical image processing and visualisation. NeuroImage, 2019
- Andersson JLR, et al. Non-linear registration, aka spatial normalisation, FMRIB technical report TR07JA2, 2010